Castor Test Case
The goal of the following test case is to reproduce Chimere test case for the 2003 Heat Wave in Europe.
Step 1: Obtaining and Installing the Test Case
You must have already downloaded and installed Polyphemus to have this test case working. Below, [version] will stand for 1.8.1 but this test case works also with versions 1.8, 1.7.x, 1.6, 1.5, 1.4, 1.3.x or 1.2.x.
- First, you need to download TestCase-Castor-1.5.tar.bz2.
To uncompress this file, execute the following command:
$ tar xjvf TestCase-Castor-[version].tar.bz2
This create a directory TestCase-Castor/ (referred to below as ~/TestCase/), containing:- a directory raw_data/, with data necessary for preprocessing.
- a directory data/, with all data precomputed. It also contains the results of preprocessing.
- a directory config/, with all configuration files used.
- a directory results/ where the results are stored.
- a program, called sum-emissions.py, and its configuration file (sum-emissions.cfg), used to sum biogenic and anthropogenic emissions.
- a file version stating for what version of Polyphemus the test-case was made.
- The test-case also requires data from Chimere test-case:
- the meteorological file
- the emission data
- the INCA data
Step 2: Modifying the General Configuration File
The file config/general.cfg is used by all preprocessing programs and as such must be the first file you modify when performing preprocessing. Make sure to modify and use the file provided in the directory TestCase/config/ .
You should only need to replace the value of Programs to have the path to preprocesing in the last version of Polyphemus compatible with the test-case you have. The domain is defined for a simulation over Europe. Make sure that the date is 2003-07-30.
Step 3: Computing Input Data
3-a) Land Data
A python program is provided among the preprocessing programs to generate land data from Chimere raw data. Files LANDPAR and LANDUSE_CONT3 from Chimere V200606B are necessary to generate land data. They have been included in the archive.
$ python ~/Polyphemus-[version]/preprocessing/ground/ground-castor.py \ config/ground-castor.cfg
This creates two files (LUC.bin and Roughness.bin) in data/ground/.
3-b) Meteorological Data
Download the meteorological file for Chimere test-case and put it in raw_data, then extract it.
$ cd raw_data $ wget http://euler.lmd.polytechnique.fr/chimere/downloads/MMOUT_EUR2_20030730_20030803.gz $ gunzip MMOUT_EUR2_20030730_20030803.gz
After you have done so, execute MM5-meteo-castor to process the MM5 file you have downloaded.
$ ~/Polyphemus-[version]/preprocessing/meteo/MM5-meteo-castor \
config/general.cfg config/MM5-meteo-castor.cfg 2003-07-30 5d2h
The output on screen will be:
Reading configuration... done. Memory allocation for grids... done. Memory allocation for output data fields... done. Conversion from sigma levels to altitudes... done. Converting from latlon to MM5 indices... done. Computing pressure... done. Computing surface pressure... done. Wind rotation... done. Horizontal interpolations... done. Vertical diffusion... done. Computing attenuation... done. Vertical averages... done. Writing data... done.
This creates 18 binary files in data/meteo/.
3-c) Anthropogenic Emissions
We generate anthropogenic emissions using emission data from Chimere test-case. Download the raw data from Chimere website and put it in raw_data:
$ cd raw_data $ wget http://euler.lmd.polytechnique.fr/chimere/downloads/AemiCONT3-200311.tar.gz $ tar xzvf AemiCONT3-200311.tar.gz
This creates a directory raw_data/AemiCONT3-200311/.
Launch the generation of emissions with the following command line:
$ ~/Polyphemus-[version]/utils/call_dates \
~/Polyphemus-[version]/preprocessing/emissions/chimere_to_castor \
config/general.cfg config/chimere_to_castor.cfg 20030730 6
Here the command line has been divided in two by "\" for clarity reason but it should be typed as one line.
We use utility program call_dates because chimere_to_castor can only be launched for one day at a time.
The output for the first day will be:
nice time ~/Polyphemus-[version]/preprocessing/emissions/chimere_to_castor \
config/general.cfg config/chimere_to_castor.cfg 20030730
Reading configuration... done.
Reading input emissions... done.
Converting to Castor and Polair3D emissions...
+ PPM_big
+ PPM_coa
+ PPM_fin
+ NO
+ NO2
+ HONO
+ CO
+ SO2
+ NH3
+ CH4
+ C2H6
+ NC4H10
+ C2H4
+ C3H6
+ APINEN
+ C5H8
+ OXYL
+ HCHO
+ CH3CHO
+ CH3COE
done.
4.80user 0.97system 0:19.73elapsed 29%CPU (0avgtext+0avgdata 0maxresident)k
0inputs+0outputs (2major+4756minor)pagefaults 0swaps
Again, the first line has been divided with "\" for clarity reasons. The program creates 20 binary files in data/emissions/.
3-d) Biogenic Emissions
Biogenic emissions are generated from meteorological data, using program bio-castor.
Launch the program with:
$ ~/Polyphemus-[version]/preprocessing/bio/bio-castor config/general.cfg \
config/bio-castor.cfg 2003-07-30 5d2h
The output on screen will be:
Reading configuration... Reading meteorological data... done. Computing biogenic emissions... done. Writing output emissions... done.
This creates three binary files in data/bio: Isoprene.bin, NO.bin and Terpenes.bin.
3-e) Summing Emissions
Anthropogenic and biogenic emissions have to be summed. They can be generated for different length of time, which is why a script has been provided to perform the sum. Launch it with:
$ python sum-emissions.py sum-emissions.cfg
The output on screen will be:
Summing anthropogenic and biogenic emissions. Anthropogenic species: NO. Biogenic species: NO. Computing total emissions for NO. Anthropogenic species: C5H8. Biogenic species: Isoprene. Computing total emissions for C5H8. Anthropogenic species: APINEN. Biogenic species: Terpenes. Computing total emissions for APINEN.
This creates three binary files in data/emissions: APINEN-total.bin, C5H8-total.bin and NO-total.bin.
3-f) Deposition Velocities
Deposition velocities using Emberson parameterization are computed with program dep-emberson. Launch it with:
$ ~/Polyphemus-[version]/preprocessing/dep/dep-emberson config/general.cfg \
config/dep-emberson.cfg 2003-07-30 5d2h
The output on screen will be:
Reading configuration files... done. Memory allocation for data fields... done. Extracting input data... done. Computing deposition velocities... done. Writing output data... done.
This computes deposition velocities for 23 species.
3-g) Boundary Conditions
Download and put in raw_data/ the INCA files from Chimere test-case.
$ cd raw_data $ wget http://euler.lmd.polytechnique.fr/chimere/downloads/INCA-200501.tar.gz $ tar xzvf INCA-200501.tar.gz
This creates a data directory named raw_data/INCA.
First you need to modify the configuration file config/bc-inca.cfg. Indeed you have to put the number of time steps for which you want boundary conditions generated. As INCA are monthly files, you need only to generate the boundary conditions in July for two days, that is to say 48 hours. The configuration file will be:
# Configuration file for inca boundary conditions. [bc_input_domain] x_min = -180. Delta_x = 3.75 Nx = 96 y_min = -90. Delta_y = 2.5 Ny = 73 Nz = 19 # Input species. Ns = 14 Species: <Programs>/bc/species_inca.dat [bc_files] Nt = 48 Directory_bc: <Directory_computed_fields>/boundary_conditions/
Then launch computation of the boundary conditions with:
$ ~/Polyphemus-[version]/preprocessing/bc/bc-inca config/general.cfg \
config/bc-inca.cfg raw_data/INCA/INCA.07
The output on screen will be:
Memory allocation for data fields... done
Reads file... done
Input data processing... done
Species:
O3 ... done
NO ... done
NO2 ... done
HNO3 ... done
PAN ... done
H2O2 ... done
CO ... done
CH4 ... done
HCHO ... done
C2H6 ... done
NC4H10 ... done
C2H4 ... done
C3H6 ... done
OXYL ... done
As the simulation is set in July and August 2003, launch the program again using INCA.08 this time and 78 (hourly) time steps in August.
You will obtain boundary conditions for 14 species.
Step 4: Launching the Simulation
4-a) Modifying the Configuration Files
You should check and modify config/castor.cfg if necessary. You have to check the paths (in particular check that the data and saver file are config/castor-data.cfg and config/castor-saver.cfg) and to make sure that the date for the simulation is 2003-07-30 (date from which the preprocessing starts).
Then check the paths and dates in config/data.cfg. In particular, if the dates in any section are not right, you can have an error message:
ERROR! An input/output operation failed in FormatBinary<T>:: Read(ifstream& FileStream, Array<TA, N>& A). Unable to read 42900 byte(s). The input stream is empty.
Indeed, input data can be computed for several days, so the program will discard the data for the days between Date_min in a section of polair3d-data and Date_min for the simulation.
Also remember that volume emissions given for NO, APINEN and C5H8 are the sum of anthropogenic and biogenic emissions.
4-b) Simulation
Launch the simulation with:
$ ~/Polyphemus-[version]/processing/castor config/castor.cfg
The output on screen will be:
Current date: 2003-07-30 00:00 Current date: 2003-07-30 00:10 Current date: 2003-07-30 00:20 Current date: 2003-07-30 00:30 Current date: 2003-07-30 00:40 Current date: 2003-07-30 00:50 Current date: 2003-07-30 01:00 Current date: 2003-07-30 01:10 Current date: 2003-07-30 01:20 Current date: 2003-07-30 01:30 Current date: 2003-07-30 01:40 Current date: 2003-07-30 01:50 [...] Current date: 2003-08-03 23:10 Current date: 2003-08-03 23:20 Current date: 2003-08-03 23:30 Current date: 2003-08-03 23:40 Current date: 2003-08-03 23:50
4-c) Checking your results
In order to check your results, type:
$ ~/Polyphemus-[version]/utils/get_info_float results/O3.bin
You should get something like:
Minimum: 0.000103238 Maximum: 134.057 Mean: 52.8842
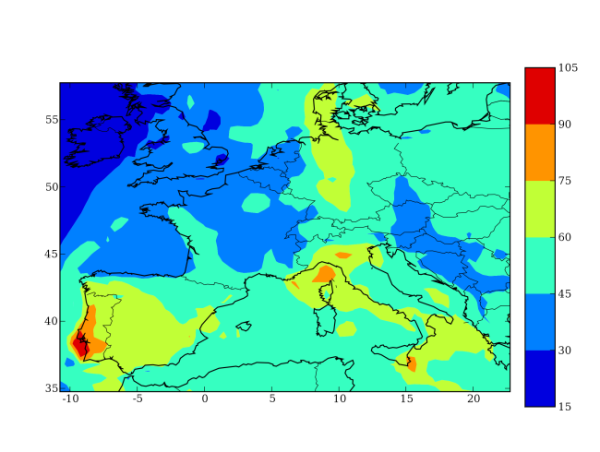
Step 5: Visualizing results
To visualize results you have to put the path to Polyphemus/include in your PYTHONPATH. For instance, for Bash users add export PYTHONPATH=$PYTHONPATH:~/Polyphemus/include to your .bashrc.
Then go the directory results/ and launch IPython.
$ cd results/
$ ipython
>>> from atmopy.display import *
>>> m = getm('disp.cfg')
>>> d = getd('disp.cfg')
>>> dispcf(m, d[40, 0]) The output figure will be:


 Support
Support